Finding Objects with Cellpose Models
Note: Segmentation using Deep Learning requires the Deep Learning extension to the 2D Automated Analysis module. The Image-Pro Neural Engine must be installed. Installing the Image-Pro Neural Engine
Image-Pro supports object segmentation using Cellpose Models. Cellpose models perform instance segmentation (they attempt to identify each example or 'instance' of your objects of interest). The Cellpose algorithm was designed as a 'generalist algorithm for cellular segmentation' by Carsen Stringer, Michalis Michaelos and Marius Pachitariu of the Janelia Research Campus (Stringer et al. 2021). The algorithm is extremely effective at segmenting the cellular objects for which it was designed, as well as a wide range of non-cellular objects of interest in materials analysis.
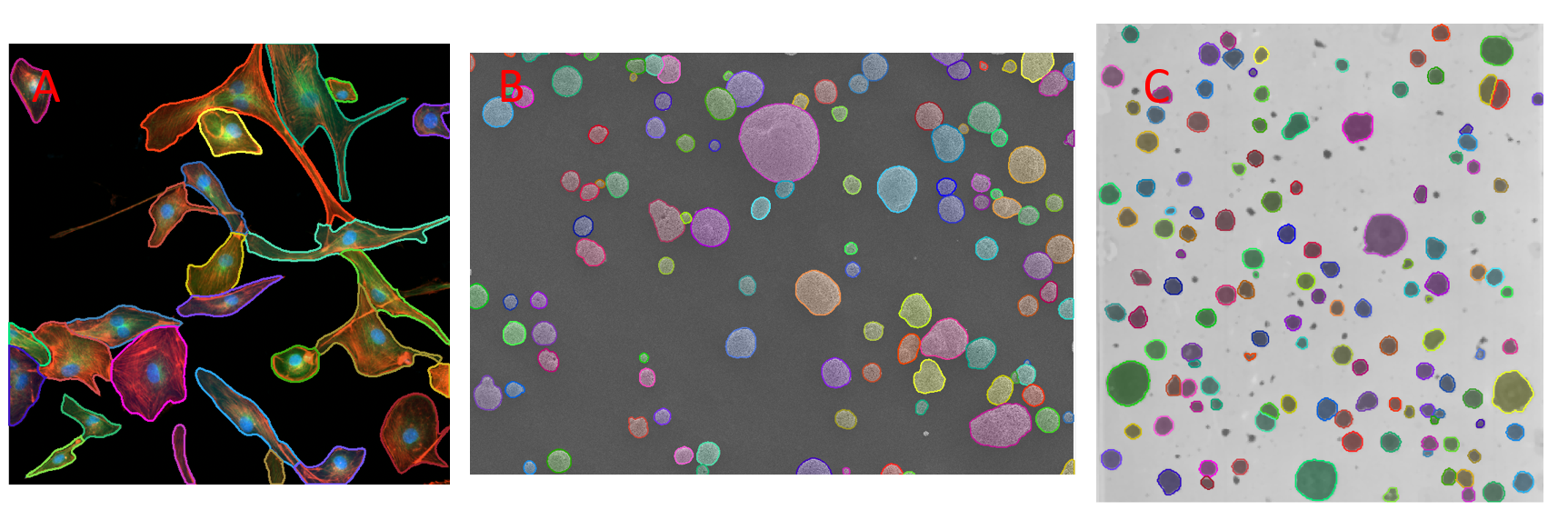
Figure showing object segmentation using Cellpose models. A). Composite fluorescence image of bovine pulmonary epithelial cells B) SEM image of particles C) Bright-field image of particles.
To segment objects using Cellpose models in the active image select the AI button from the Segment tool group on the Count/Size ribbon. The AI Deep Learning Prediction panel will open.
Perform the following actions within the AI Deep Learning Prediction panel:
-
Load Model.
-
Click the Load button. The Open Model for Prediction dialog opens.
-
Select Cellpose Instance Segmentation from the Architecture drop down to show only Cellpose models.
-
Use the example images and description to help you select a Cellpose model that's appropriate for your objects of interests and click Open.
-
-
Configure Classes
-
Name the class and set the color and shape of predicted objects.
-
-
Select Targets.
-
Select the channel that contains your objects of interest as the Segmentation Channel.
-
Optional. Select the channel that contains secondary identifying objects as the Seed Channel
Note: Setting a Seed Channel only makes sense if the selected model was trained with a secondary channel. For example, the Fluorescent Cells model was trained with secondary channel that contains labeled nuclei.
-
-
Set Model Options.
-
Check the Object Diameter by comparing the size guide to your objects of interest. If the size guide does not represent your objects of interest either:
-
Manually set the size by entering a value in the Object Diameter Field
-
Manually change the size guide by selecting and dragging the control points
-
Click Estimate (this option is only displayed for models with a trained size model) and check the size guide again.
-
-
We recommend that the default settings for the remaining model options are initially applied. You can refine these options as described below following your prediction.
-
- Predict and Count.
-
Select the Show Output check box to automatically open the Deep Learning Output panel while prediction takes place. The Deep Learning Output panel will display information and feedback from the Cellpose model during prediction.
-
Select the Apply Filters/Splits check box if you wish to Count objects in the image with ranges and automatic splits applied. This is not recommended for instance segmentation with Cellpose models as it can become difficult to determine exactly what was predicted by the model.
-
Click Count. Objects of interest that are found in your image will be displayed as measurement objects.
-
Use the Show/Hide button to toggle the display of objects on the image and to determine if you are happy that the measured objects are accurate.
-
Refining Cellpose Prediction
If you fail to find the expected number of objects when making a Cellpose prediction, there are several adjustments that can be made to improve the prediction in Step 4 of the AI Deep Learning Prediction panel - Set Model Options.
Channel Selection
Both a Segmentation and an Optional Channel can be set in the Select Targets section of the AI Deep Learning Prediction panel. In many cases, highly accurate segmentation can be generated without setting the Optional Channel. In cases where the model was trained with an optional second channel, and where your data set contains this second channel, you can greatly improve the segmentation by correctly selecting the second Optional Channel. The Fluorescent Cells model was trained with the segmentation channel set to the channel with the cytoplasm label, and the second Optional Channel set to the channel with the nuclear label.
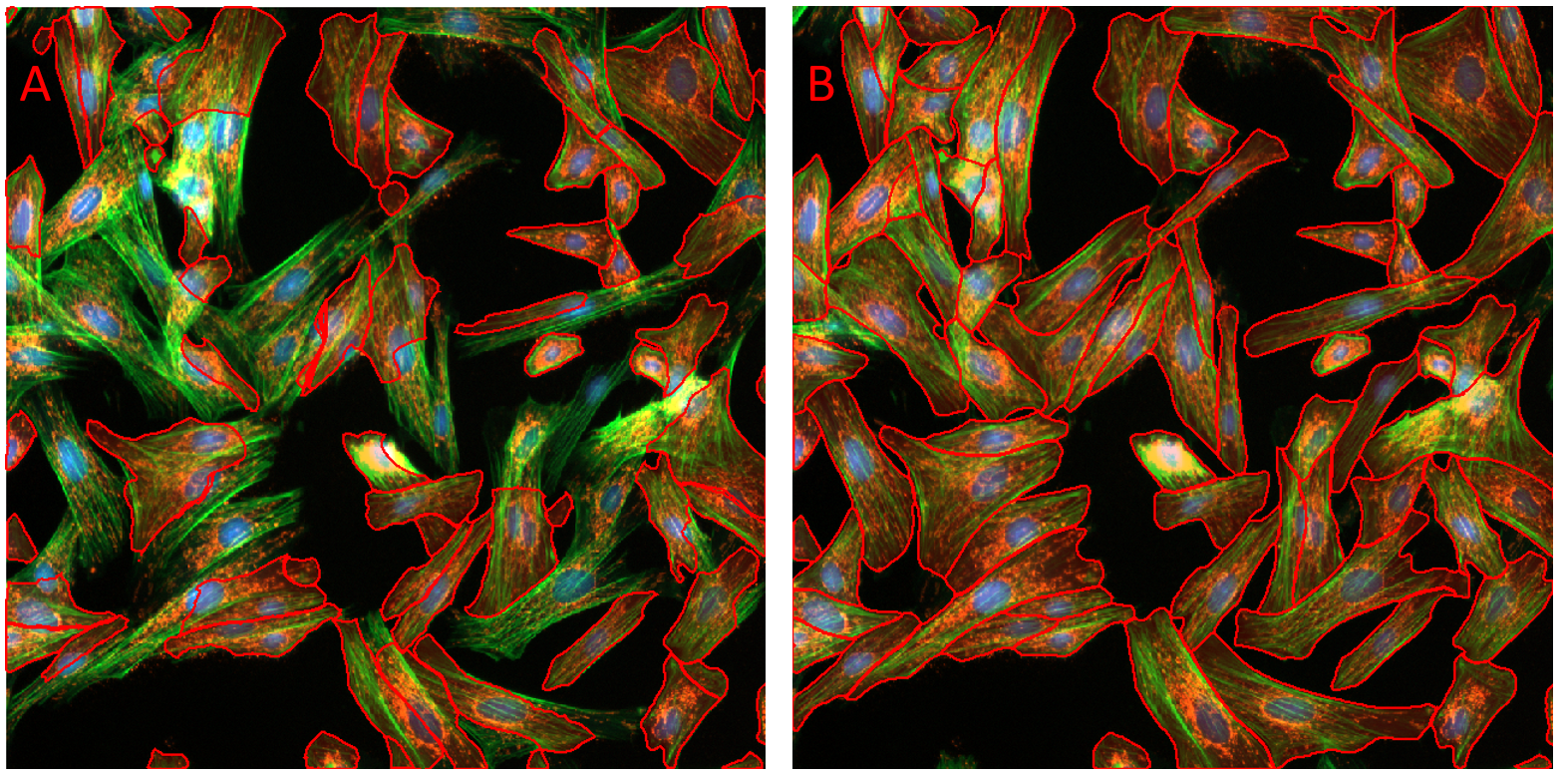
Figure showing a composite fluorescence image of bovine pulmonary epithelial cells. Objects were segmented using the Fluorescent Cells model using default setting. In panel A no Optional Channel was set. In panel B the channel containing the nucleus label was set as the Optional Channel. Note the great improvement in segmentation with correct Optional Channel set.
Object Diameter
Accurate prediction by CellPose models is highly dependent on the accuracy of the Object Diameter setting.
Note: If your image is calibrated, the Object Diameter is shown in calibrated units, the diameter is shown in pixels for uncalibrated images.
If supported by your model, clicking Estimate will automatically populate this field. You should always check, and if necessary edit estimated Object Diameter settings.
Note: If your model does not support the Estimate function, the option is not shown and is replaced with the Default check box. When selected the Object Diameter is set to the mean diameter of objects in the model's training data. You must deselect the default check box and edit the Object Diameter if the default value does not accurately reflect the diameter of your objects of interest.
Select the Size Guide Display Visible check box to display a graphical representation of Object Diameter on the active image.
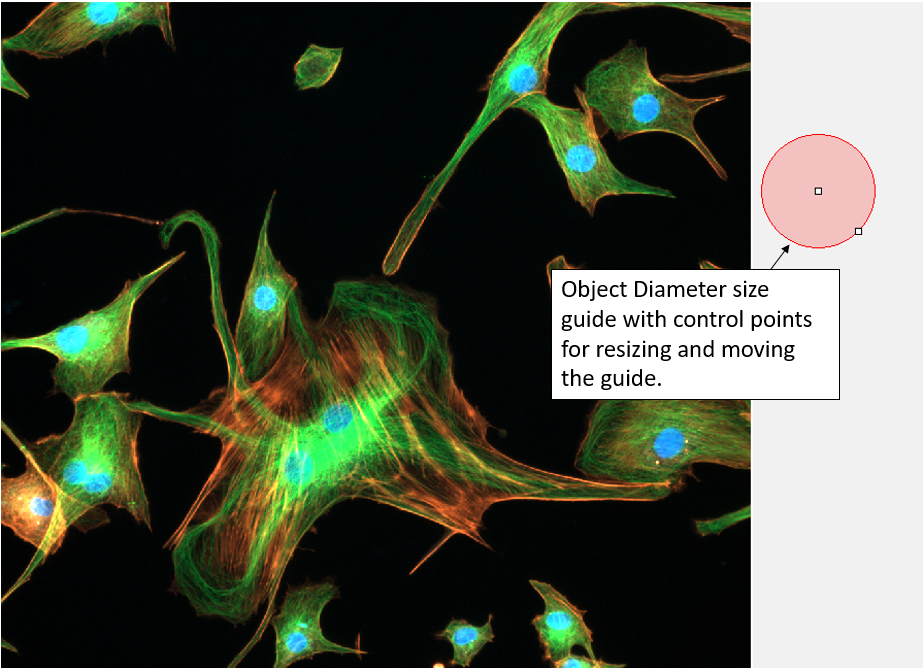
You can change the Object Diameter setting by clicking on the size guide and dragging the control points. You can also use the control points to move the guide around the active image, or to the side of the active image.
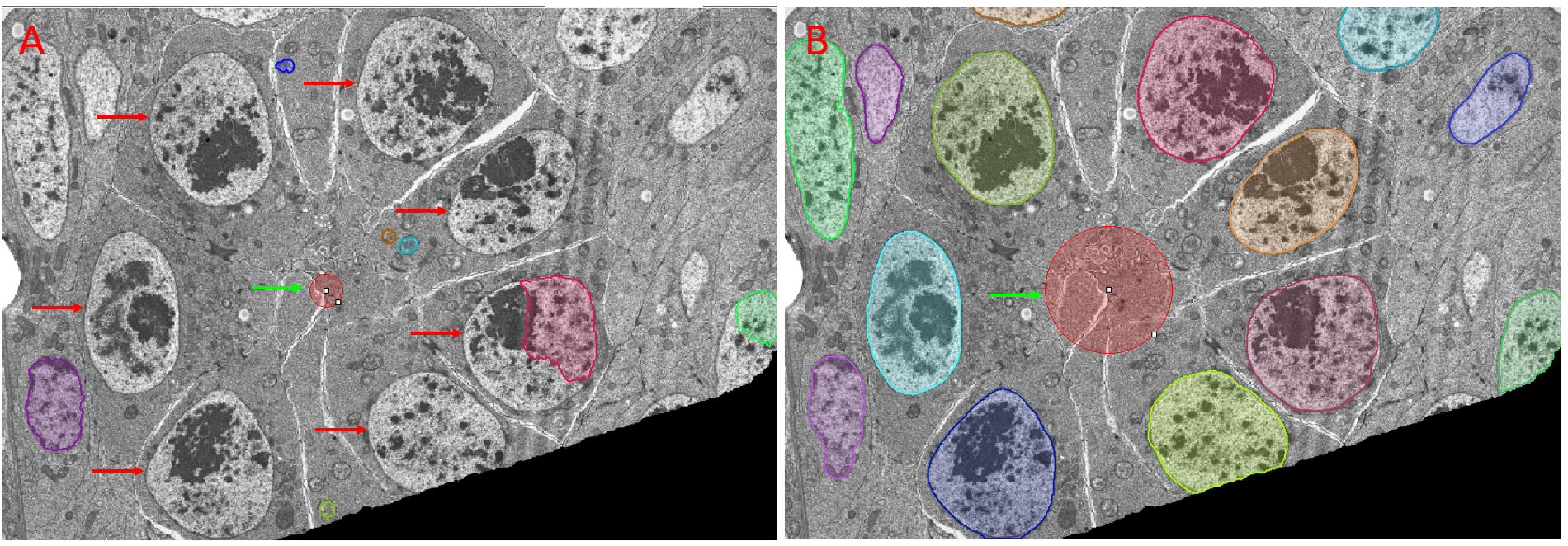
Figure of images after Prediction with a CellPose model showing the effect of the Object Diameter setting. Panel A: Objects of interest (against which the model was trained) are highlighted with red arrows. The size guide representing the Object Diameter used for prediction is highlighted by the green arrow. The failure to correctly set Object Diameter results in failure to predict the objects of interest. Panel B: The Object Diameter was set by clicking on the size guide (green arrow) and dragging the control points until the guide was within the size range of the objects of interest. All objects of interest were accurately predicted after correctly setting Object Diameter.
Strictness
The strictness setting allows you to filter objects according to the likelihood that they are genuine. Reducing this setting increases the number of objects predicted, but increases the risk that some objects will be incorrect. Increasing this setting decreases the number of objects predicted, but increases the likelihood that predicted objects will be correct. Setting the strictness to 0 results in no filter being applied and the maximum number of objects being predicted.
Note: For users familiar with Cellpose documentation, Strictness is an implementation of the Flow Threshold setting. Flow Threshold is applied as 100 - Strictness. Cellpose Settings
Probability Threshold
The probability that every pixel in the image is part of an object is predicted by the CellPose model. Pixels with a probability above this threshold are used to determine objects. You generate more or larger objects by reducing the probability threshold, and fewer or smaller objects by increasing the threshold.
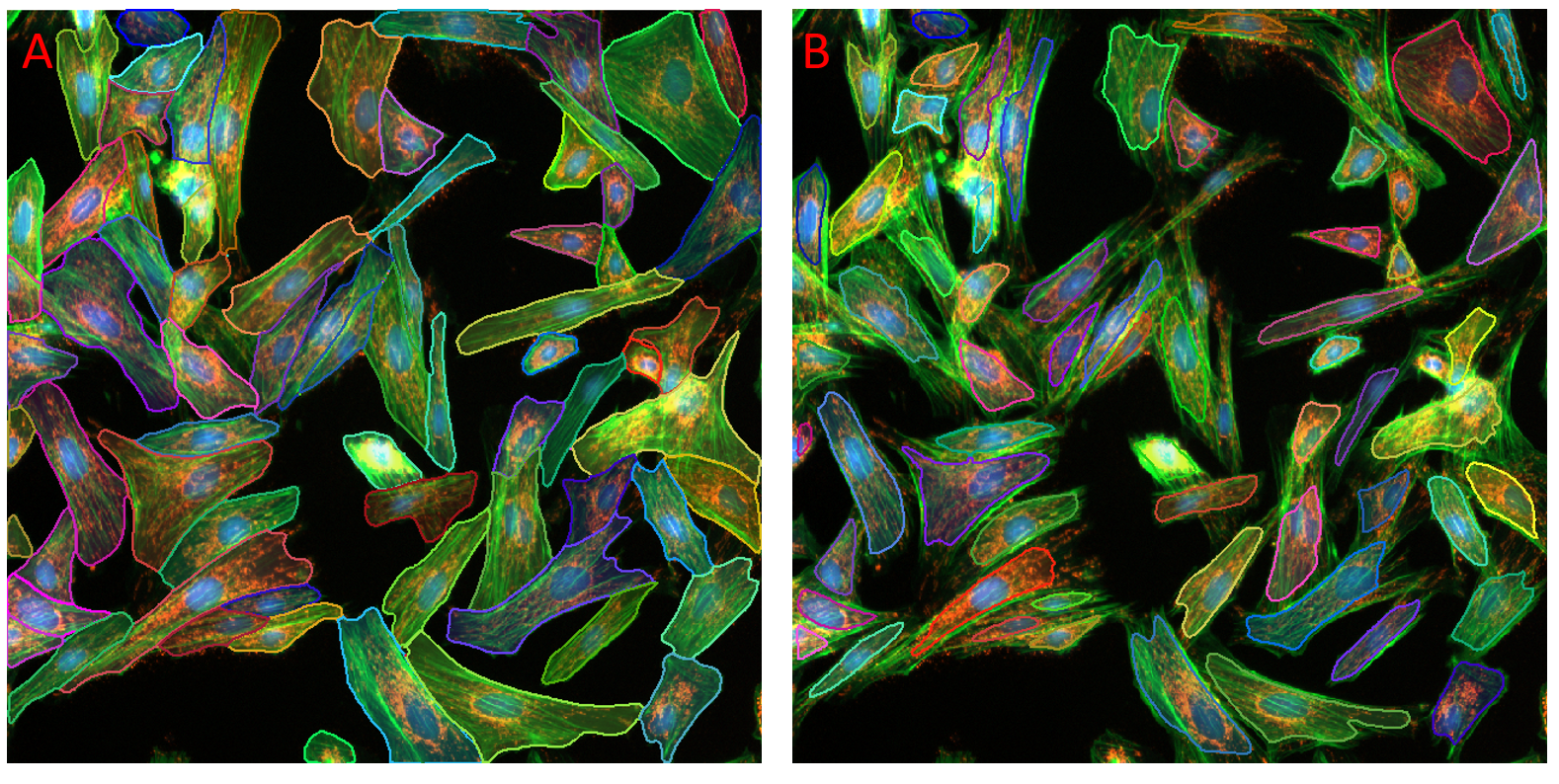
Figure showing a composite fluorescence image of bovine pulmonary epithelial cells. Cells were found using the Fluorescent Cells model. In panel A the default probability threshold of 60% was applied. In panel B the probability threshold was increased to 90%. Fewer and smaller objects are found in panel B.
Sparse Mode
By default this setting is off. When sparse mode is off a new image for analysis is calculated in which the data is normalized in which the 1st percentile of image intensity is set to 0.0 and the 99th percentile of image intensities is set to 1.0 in each channel.
In cases where the ratio of objects to background is very low, and an unexpectedly low number of objects are found, sparse mode should be enabled. With spare mode enabled, the prediction is done using the Image's current display range.
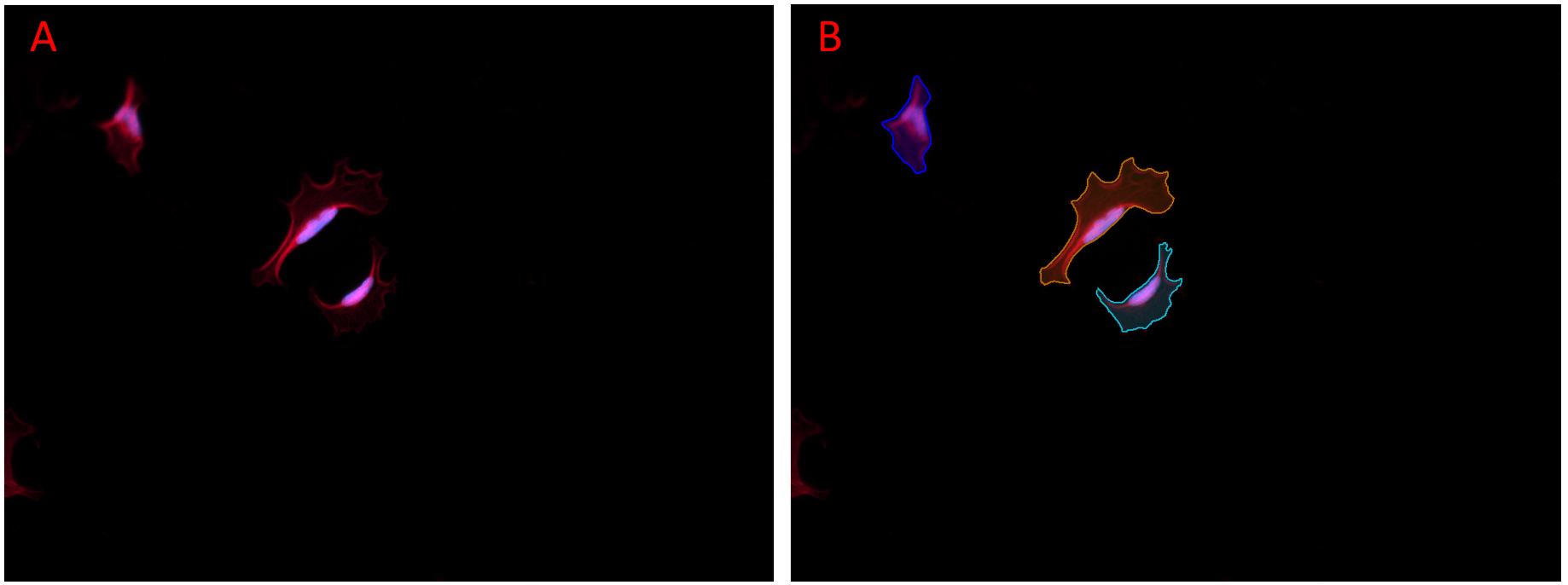
Figure showing an image with a low ratio of objects to background that benefits from Sparse Mode. In Panel A, prediction was done with Sparse Mode disabled, no objects were found. In Panel B, Sparse Mode was enabled and all objects in the image were found. All other settings were identical.
Image Areas in a Different Intensity Range
Inclusion of areas of images in a different intensity range to both the objects of interest and the background can degrade Cellpose predictions. Such areas can affect the normalization of the data (which Cellpose does in the background) in a way that makes target objects less visible to the model. When possible such areas should be excluded using a region of interest.
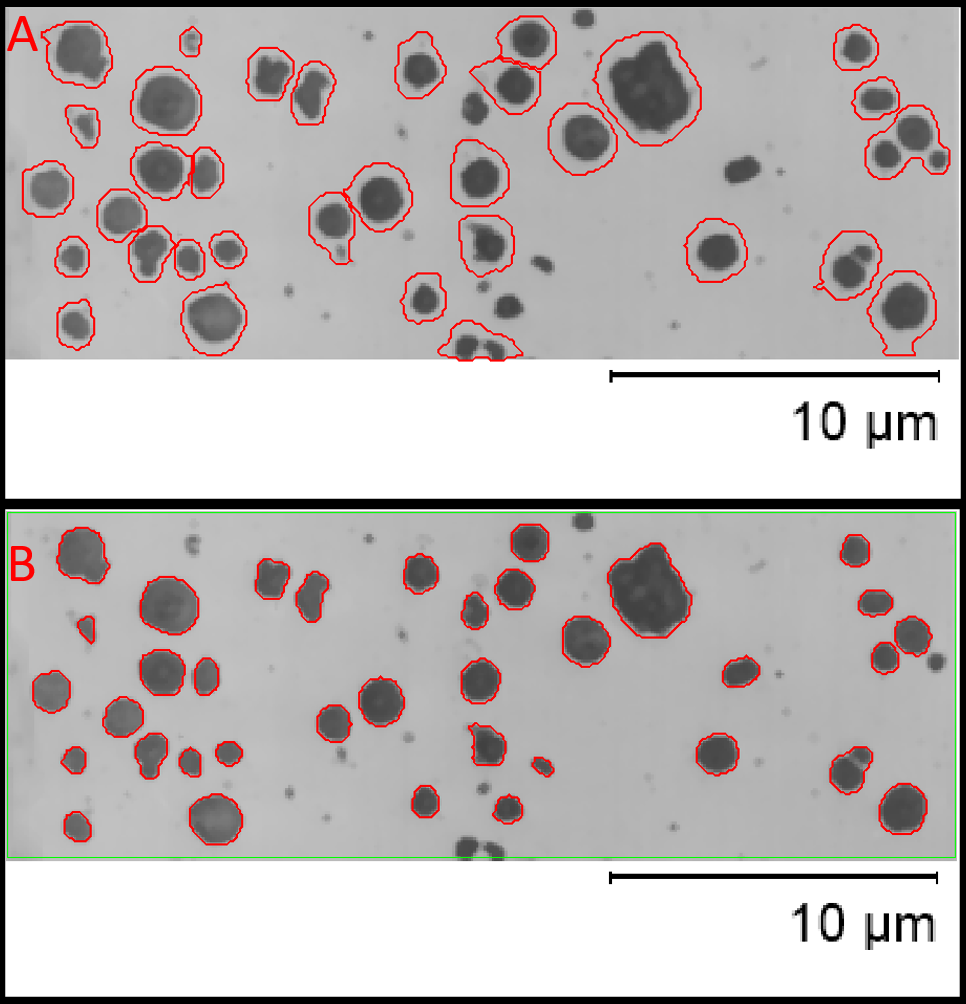
Figure illustrating the effect of image areas in a different intensity range to the area of interest, in this case an information area with a scale bar in which pixels are set to the maximum gray level of the image. In Panel A this area in included in the prediction, note the poor segmentation of objects, failure to discriminate pairs and failure to segment some objects. This area is excluded from the segmentation with a region of interest (green) in panel B, this generates greatly improved accuracy.
If you are unable to predict your objects of interest having exhausted all of the refinement steps, you can refine your model with further training.
Click Continue Training to open the model in the AI Deep Learning Training Panel.
Locked models can not be further trained. Click Clone Model to open the Clone Model dialog. The cloned model can be opened in the trainer and improved with additional training while keeping the current model unchanged.
Saving Deep Learning Segmentation Settings
It's convenient to save all of your deep learning settings including your model, model options, range settings, and split settings as a single settings file. To do this, click Save in the Measurements tool group on the Count/Size ribbon. A Save Measurements Options dialog opens. Name and save your measurements options (.iqo) file.
Restoring Saved Deep Learning Segmentation Settings
Click Load in the Measurements tool group on the Count/Size ribbon. A Load Measurements Options dialog opens. Navigate to your saved measurements options (.iqo) file and click Open. Your saved settings are restored.
Bibliography
Stringer, C., Wang, T., Michaelos, M. & Pachitariu, M. (2021) Cellpose: a generalist algorithm for cellular segmentation. Nat Methods 18, 100–106. https://doi.org/10.1038/s41592-020-01018-x
Learn more about all features in Count/Size